AmeriSpeak® is a research panel where members get rewards when they share their opinions. Members represent their communities when they answer surveys online, through the AmeriSpeak App, or by phone. Membership on the AmeriSpeak Panel is by invitation only. Policymakers and business leaders use AmeriSpeak survey results to make decisions that impact our lives. Tell us your thoughts on issues important to you, your everyday life, and topics in the news such as health care, finance, education, technology, and society.
Commission launches call to create the European Digital Media Observatory The European Commission has published a call for tenders to create the first core service of a digital platform to help fighting disinformation in Europe. The European Digital Media Observatory will serve as a hub for fact-checkers, academics and researchers to collaborate with each other and actively link with media organisations and media literacy experts, and provide support to policy makers. The call for tenders opened on 1 October and will run until 16 December 2019.
ASRC GOES 7:00 – 7:00
- Expense Report!
- Call Erikson!
- Dissertation
- Change safe to low risk
- Tweaking the Research Design chapter
- Evolver
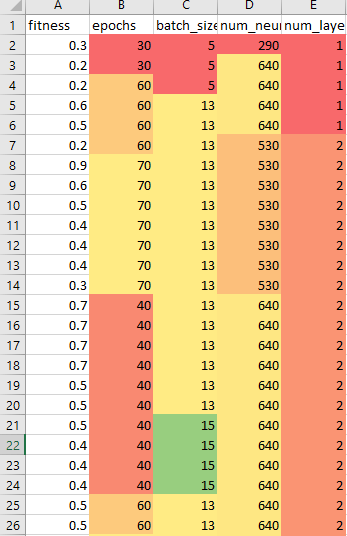
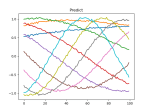
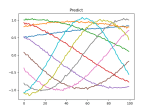
- See if the run broke or completed this weekend – IT restarted the machine. Restarted and let it cook. I seem to have fixed the GPU bug, since it’s been running all day. It’s 10,000 models!
- Look into splitting up and running on AWS
- Rather than explicitly gathering ten runs each time for each genome, I could hash the runs by the genome parameters. More successful genomes will be run more often.
- Implies a BaseEvolver, LazyEvolver, and RigerousEvolver class
- Neural Network Based Optimal Control: Resilience to Missed Thrust Events for Long Duration Transfers
- (pdf) A growing number of spacecraft are adopting new and more efficient forms of in-space propulsion. One shared characteristic of these high efficiency propulsion techniques is their limited thrust capabilities. This requires the spacecraft to thrust continuously for long periods of time, making them susceptible to potential missed thrust events. This work demonstrates how neural networks can autonomously correct for missed thrust events during a long duration low-thrust transfer trajectory. The research applies and tests the developed method to autonomously correct a Mars return trajectory. Additionally, methods for improving the response of neural networks to missed thrust events are presented and further investigated.
- Ping Will for Thursday rather than Wednesday done – it seems to be a case where the first entry is being duplicated
- Arpita’s presentation:
- Information Extraction from unstructured text
- logfile analysis
- Why is the F1 score so low on open coding with human tagging?
- Annotation generation slide is not clear





You must be logged in to post a comment.